
| Home |
| Paper |
| Paper Figures |
| Supplemental Data |
| PeakFinder |
| Protocols |
| Links |
| Feedback |
| PeakFinder Program |
|---|

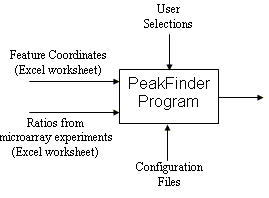
The PeakFinder program was developed to find cohesin binding sites represented by the peaks in yeast chromatin immunoprecipitation (ChIP) microarray data, but can be applied to plot any measurement against a parameter such as genome coordinate, to interactively analyze the measurement plot, and to annotate the peaks on the basis of local properties of the curve.
 |
 |
|---|
Software Requirements. PeakFinder was developed and tested under Windows 2000 (but should work under Windows 98 or later). PeakFinder works best with a high-color or true-color display monitor, and requires a resolution of 1280-by-1024 or larger. PeakFinder provides sample data to exercise the various program options.
Installation. The Installshield installation program installs PeakFinder in its own C:\PeakFinder directory. Start the program from the desktop icon:
![]()
Or, use: Start | Programs | PeakFinder.
In addition to the program, the C:\PeakFinder directory contains sample Excel spreadsheets, Coordinates.xls and Ratios.xls. The C:\PeakFinder\Yeast directory has yeast (S. cerevsiae) nucleotide files in FASTA format, one for each chromosome. The C:\PeakFinder\Documenation directory contains the Quick Start Guide and User's Guide in PDF format.
Download
Peak finder is written in Delphi, runs on a Windows platform and is distributed under the GNU General Public License.- PeakFinder Installshield Installation Program (includes documentation listed below)
- PeakFinder Quick Start Guide
- PeakFinder User's Guide
Source Code
The program has only been compiled and tested using Delphi 6 with the Excel2000 unit. I have never resolved issues compiling the code in Delphi 7 or later and with the ExcelXP unit. Unfortunately, I do not plan to update the source code for these new versions of Delphi.
The TGIFImage component, originally written by Anders Melander, is needed to compile the program. This component can be obtained from Finn Tolderlund's site. Install this component with these selections: Component | Install component | gifimage.pas | Open | OK | Yes | OK.